GeoGraph: Geometric and Graph-based Ensemble Descriptors for Intrinsically Disordered Proteins
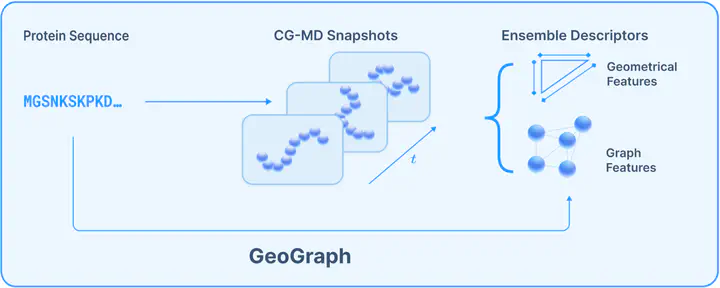 Image by Quinn et al. on Arxiv
Image by Quinn et al. on ArxivAbstract
While deep learning has revolutionized the prediction of rigid protein structures, modelling the conformational ensembles of Intrinsically Disordered Proteins (IDPs) remains a key frontier. Current AI paradigms present a trade-off. Protein Language Models (PLMs) capture evolutionary statistics but lack explicit physical grounding, while generative models trained to model full ensembles are computationally expensive. In this work we critically assess these limits and propose a path forward. We introduce GeoGraph, a simulation-informed surrogate trained to predict ensemble-averaged statistics of residue-residue contact-map topology directly from sequence. By featurizing coarse-grained molecular dynamics simulations into residue- and sequence-level graph descriptors, we create a robust and information-rich learning target. Our evaluation demonstrates that this approach yields representations that are more predictive of key biophysical properties than existing methods.