 Image by Tarasov et al. on BioRxiv
Image by Tarasov et al. on BioRxivAbstract
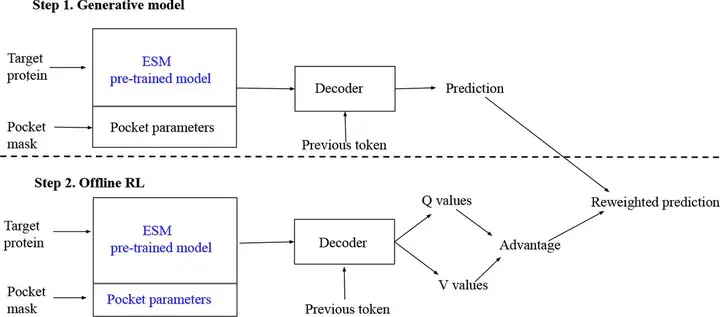
Offline Reinforcement Learning (RL) offers a compelling avenue for solving RL problems without the need for interactions with an environment, which may be expensive or unsafe. While online RL methods have found success in various domains, such as de novo Structure-Based Drug Discovery (SBDD), they struggle when it comes to optimizing essential properties derived from protein-ligand docking. The high computational cost associated with the docking process makes it impractical for online RL, which typically requires hundreds of thousands of interactions during learning. In this study, we propose the application of offline RL to address the bottleneck posed by the docking process, leveraging RL’s capability to optimize non-differentiable properties. Our preliminary investigation focuses on using offline RL to conditionally generate drugs with improved docking and chemical properties.