 Image by Zhang et al. on BioRxiv
Image by Zhang et al. on BioRxivAbstract
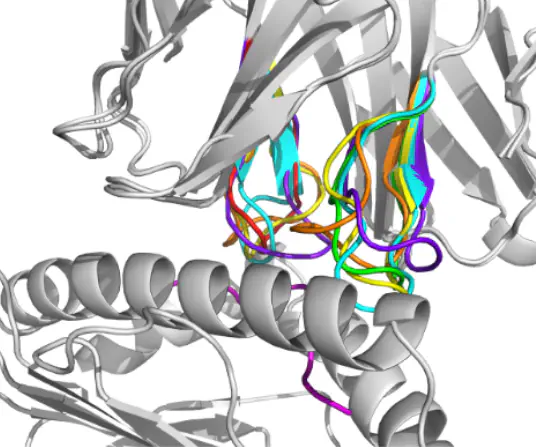
Protein structure prediction field has been revolutionised by deep learning with protein folding models such as AlphaFold 2 and ESMFold. These models enable rapid in silico prediction and have been integrated into de novo protein design and protein-protein interaction (PPI) prediction. However, biologically relevant features dependent on conformational distributions cannot be estimated with these models. Diffusion models, a novel class of generative models, have been developed to learn conformational distributions and applied to de novo protein design. Limited work has been done on protein structure inpainting, where a masked section is recovered by simultaneously conditioning on its sequence and the rest of the structure. In this work, we propose FrameDiff inPainTing (FrameDiPT), a generalised model for protein inpainting. This is important for T-cells given the hyper-variability of the complementarity determining region (CDR) loops. We evaluated the model on CDR loop design for T-cell receptors and achieved comparable prediction accuracy to ProteinGenerator and RFdiffusion with limited training data and learnable parameters. Different from deterministic structure prediction models, FrameDiPT captures the conformational distribution at different regions and binding states, highlighting a key advantage of generative models.