Generative Model for Small Molecules with Latent Space RL Fine-Tuning to Protein Targets
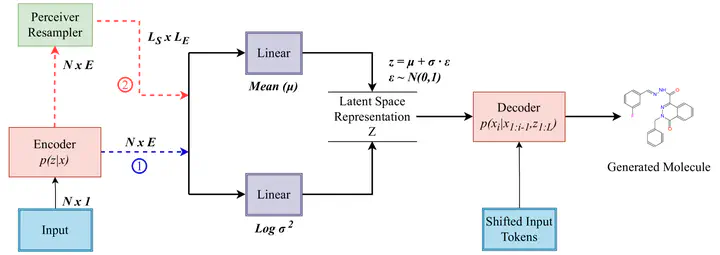 Image by Sob et al. on Arxiv
Image by Sob et al. on ArxivAbstract
A specific challenge with deep learning approaches for molecule generation is generating both syntactically valid and chemically plausible molecular string representations. To address this, we propose a novel generative latent-variable transformer model for small molecules that leverages a recently proposed molecular string representation called SAFE. We introduce a modification to SAFE to reduce the number of invalid fragmented molecules generated during training and use this to train our model. Our experiments show that our model can generate novel molecules with a validity rate > 90% and a fragmentation rate < 1% by sampling from a latent space. By fine-tuning the model using reinforcement learning to improve molecular docking, we significantly increase the number of hit candidates for five specific protein targets compared to the pre-trained model, nearly doubling this number for certain targets. Additionally, our top 5% mean docking scores are comparable to the current state-of-the-art (SOTA), and we marginally outperform SOTA on three of the five targets.