Abstract
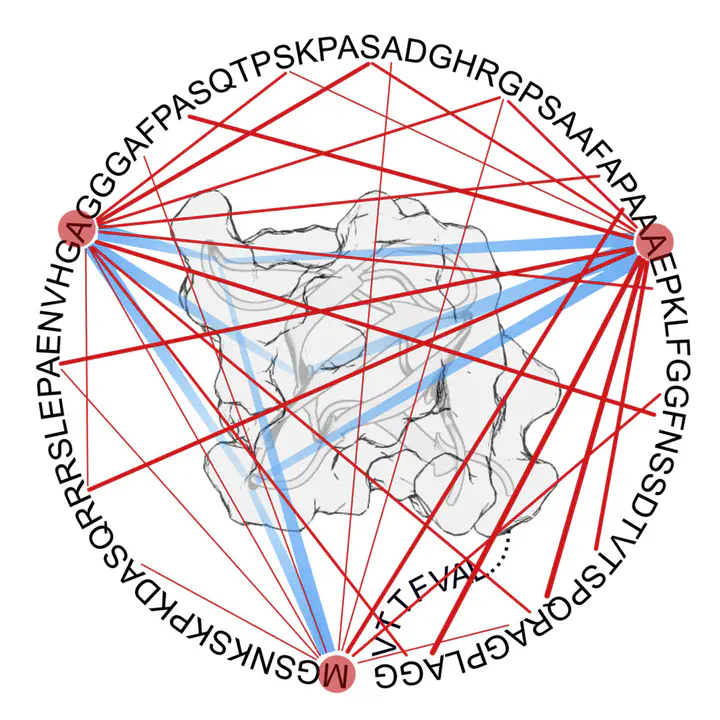
c-Src is a central player in several cellular signaling pathways. It controls important cellular processes like cellular proliferation, survival or motility. Therefore, a number of tumoral diseases have been related to abnormal c-Src activity. Among them, colorectal cancer stands out, as c-Src deregulation correlates with tumor progression and clinical outcome. This tyrosine kinase is part of a larger group of functionally and structurally related proteins termed Src Family Kinases. These proteins share the same domain architecture, a cassette formed by a catalytic domain (SH1), two reg- ulatory domains, SH2 and SH3, and a variable intrinsically disordered region (the Unique domain) that ultimately anchors to the inner face of the cellular membrane via the N-terminal SH4 domain, also disordered. The sequence and structure of the cassette are highly conserved, and thus unsurprisingly Src Family Kinases perform closely related and often overlapping functions. However, the role of intrinsically disordered regions has remained unclear, although they are known to be functionally relevant. In this work, the structural and functional relationship between the intrinsically disordered SH4 and Unique domains with the neighboring folded SH3 domain in c-Src is explored. Interactions between disordered and ordered proteins are often characterized by the formation of complexes that are specific and functional but structurally heterogeneous. Moreover, conformational plasticity is a fundamental feature for function. These assemblies are known as fuzzy complexes. Here this theoretical framework, usually applied to isolated partners, is extended to the intramolecular interface between covalently bound domains instead of isolated pairs. The concept of fuzzy binding is also used in order to describe interactions based on sets of dynamic, transient, and promiscuous contacts between ill-defined sets of interactors. In order to characterize the system, an integrative strategy using short and long range Nuclear Magnetic Resonance techniques and Small Angle X-ray Scattering is applied to several constructs containing different combinations of bound or isolated domains. It is demonstrated that the folded SH3 domain acts as a scaffold for the disordered region, which interacts in a specific manner with its partner. Both disordered domains, SH4 and Unique, are involved in the process albeit they contribute differently. Additionally, it is shown that the Unique domain is not a random coil, but contains a significant degree of pre-arrangement that is independent of the scaffold. Sequence determinants are then searched by comparison of the sequences of different Src Family Kinases. Four conserved phenylalanine residues are found and their implication in Unique domain pre-organization and Unique-SH3 domain interaction tested. All these amino acids are found to favor compaction of the intrinsically disordered region, and at the same time to perturb close contact with the scaffold. In addition, mutations in the interacting zones of the SH3 domain are also studied to test reciprocity. In all, the fuzzy complex model is proven for the SH4-Unique-SH3 system. Then, the results are extrapolated to the full-length c-Src to test its biological relevance. A coevolutionary analysis suggests that the fuzzy model may be a general feature for the whole Src Family, so the closest member of the family, Yes, is also tested experimentally. The initial results on long-range contacts suggests a similar arrangement between the scaffold and the disordered region. In all, it is suggested that plastic, fuzzy interfaces between ordered and disordered domains may be a relevant mode for the transmission of functional information within multidomain proteins. Finally, a first approach for a structural study of the c-Src fuzzy complex in a native-like lipid environment, including natural co-translational modifications, is presented. A protocol for sample preparation is developed and Dynamic Nuclear Polarization solid state NMR is shown to be an adequate tool for further analysis.